U-Mamba
Enhancing Long-range Dependency for Biomedical Image Segmentation
1Peter Munk Cardiac Centre, University Health Network, Toronto,
Canada
2Department of Laboratory Medicine and Pathobiology, University of
Toronto,
Toronto, Canada
3Vector Institute for Artificial Intelligence, Toronto,
Canada
4Department of Computer Science, University of Toronto, Toronto,
Canada
5AI Hub, University Health Network, Toronto, Canada
bowang@vectorinstitute.ai
Highlights
- We propose U-Mamba, A general-purpose segmentation network for 2D and 3D biomedical images.
- U-Mamba combines the advantage of convolutional layers and state space models, which can simultaneously capture local features and aggregate long-range dependencies.
- U-Mamba enjoys a self-configuring mechanism, allowing it to automatically adapt to various datasets without manual intervention.
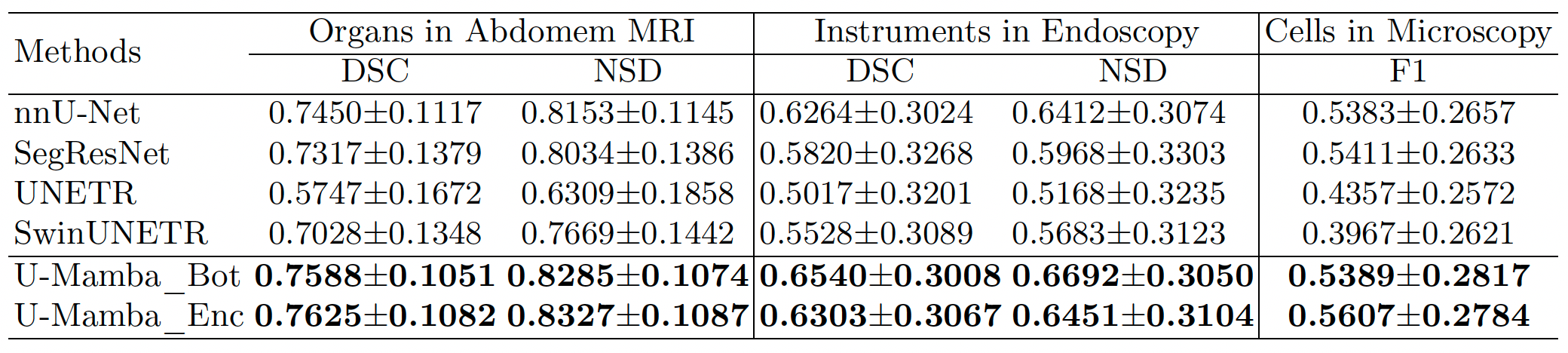
- Extensive experiments demonstrate U-Mamba achieves superior performance than both CNN- and Transformer-based networks on four diverse tasks, including the 3D abdominal organ segmentation in CT and MR images, instrument segmentation in endoscopy images, and cell segmentation in microscopy images.
- The results suggest that U-Mamba is a promising candidate for serving the next-generation biomedical image segmentation network backbone.
Network

Overview of the U-Mamba (Enc) architecture. a. U-Mamba building block contains two successive Residual blocks followed by a Mamba block to enhance long-range dependencies. b. The whole architecture of U-Mamba, which is a typical encoder-decoder framework with U-Mamba blocks in the encoder, Residual blocks in the decoder, together with skip connections.
Experiments and Results
Dataset Overview
We evaluate U-Mamba on four publicly available datasets, covering four modalities and various targets. U-Mamba inherits the self-configuring feature from nnU-Net, which can automatically generate networks for different datasets.
| Dataset | Dimension | #Training Image | #Testing Image | #Targets |
|---|---|---|---|---|
| Abdomen CT | 3D | 50 (4794 slices) | 50 (10894 slices) | 13 |
| Abdomen MRI | 3D | 60 (5615 slices) | 50 (3357 slices) | 13 |
| Endoscopy Images | 2D | 1800 | 1200 | 7 |
| Microscopy Images | 2D | 1000 | 101 | 2 |
| Configurations | Patch size | Batch size | # Stages | # Pooling per axis |
|---|---|---|---|---|
| Abdomen CT | (40, 224, 192) | 2 | 6 | (3, 3, 5) |
| 3D Abdomen MR | (48, 160, 224) | 2 | 6 | (3, 5, 5) |
| 2D Abdomen MR | (320, 320) | 30 | 7 | (6, 6) |
| Endoscopy | (384, 640) | 13 | 7 | (6, 6) |
| Microscopy | (512, 512) | 12 | 8 | (7, 7) |
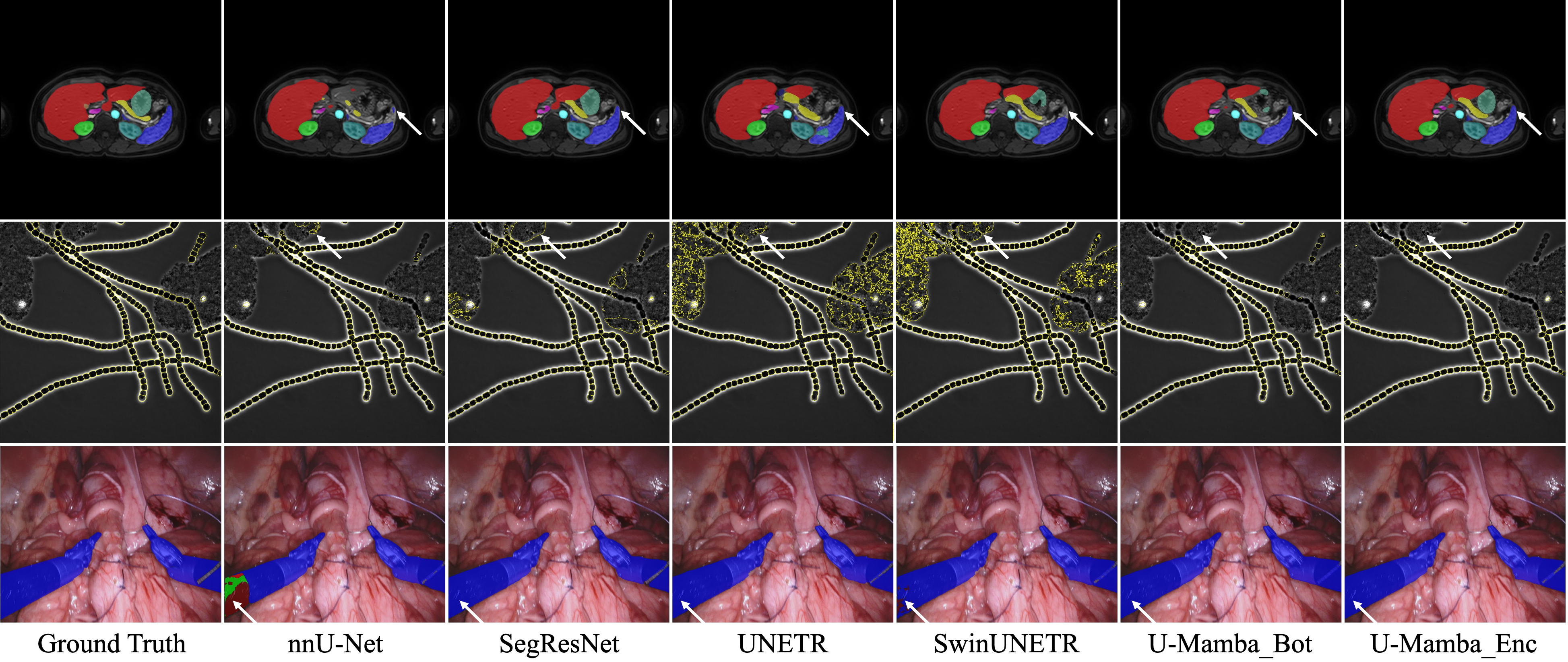
Results
Visualized results demonstrate that U-Mamba can handle complex targets with fewer segmentation outliers.


BibTeX
@article{U-Mamba,
title={U-Mamba: Enhancing Long-range Dependency for Biomedical Image Segmentation},
author={Ma, Jun and Li, Feifei and Wang, Bo},
journal={arXiv preprint arXiv:2401.04722},
year={2024}
}
This website template is borrowed from here. Thank you!